With the rise of the advanced high throughput technologies in molecular biology, the applications of bioinformatics are widely needed for the biological data analysis and management.
Our services mainly focus on three key areas:
Our core has provided in-depth data analysis services and research supports to our users in various research projects, including chromosome biology, developmental biology, infection and immunobiology, Neuroscience, Plant Biology, RNA Biology, Structural Biology, and Systems Biology, and more. Our team work closely with users to analyze, interpret, and distribute scientific discoveries derived from the analysis of high-throughput data. In addition, we also provide the high-performance computational resources (linux cluster servers) for all IMB labs to conduct their own bioinformatics analysis.
In conclusion, the objective of the IMB Bioinformatics Core is to provide all IMB researchers with up-to-date bioinformatics and computational expertise to ensure the comprehensive supports in all phases of biological researchs.
Projects Completed
Labs
Users
User Satisfaction (/5)
Experimental Design
Statistical Consultation
Software/Database Support
Genome Assembly
Genome Variant Calling
Bulk RNA-Seq
Single-Cell RNA-Seq
Methylation (BS-Seq)
ChIP-Seq and ATAC-Seq
Protein structure prediction
Functional enrichment
Custom NGS Analyses
Functional Analysis
Proteomics Analysis
Metabolomics Analysis
Protein Structural Analysis
Image Analysis
Others
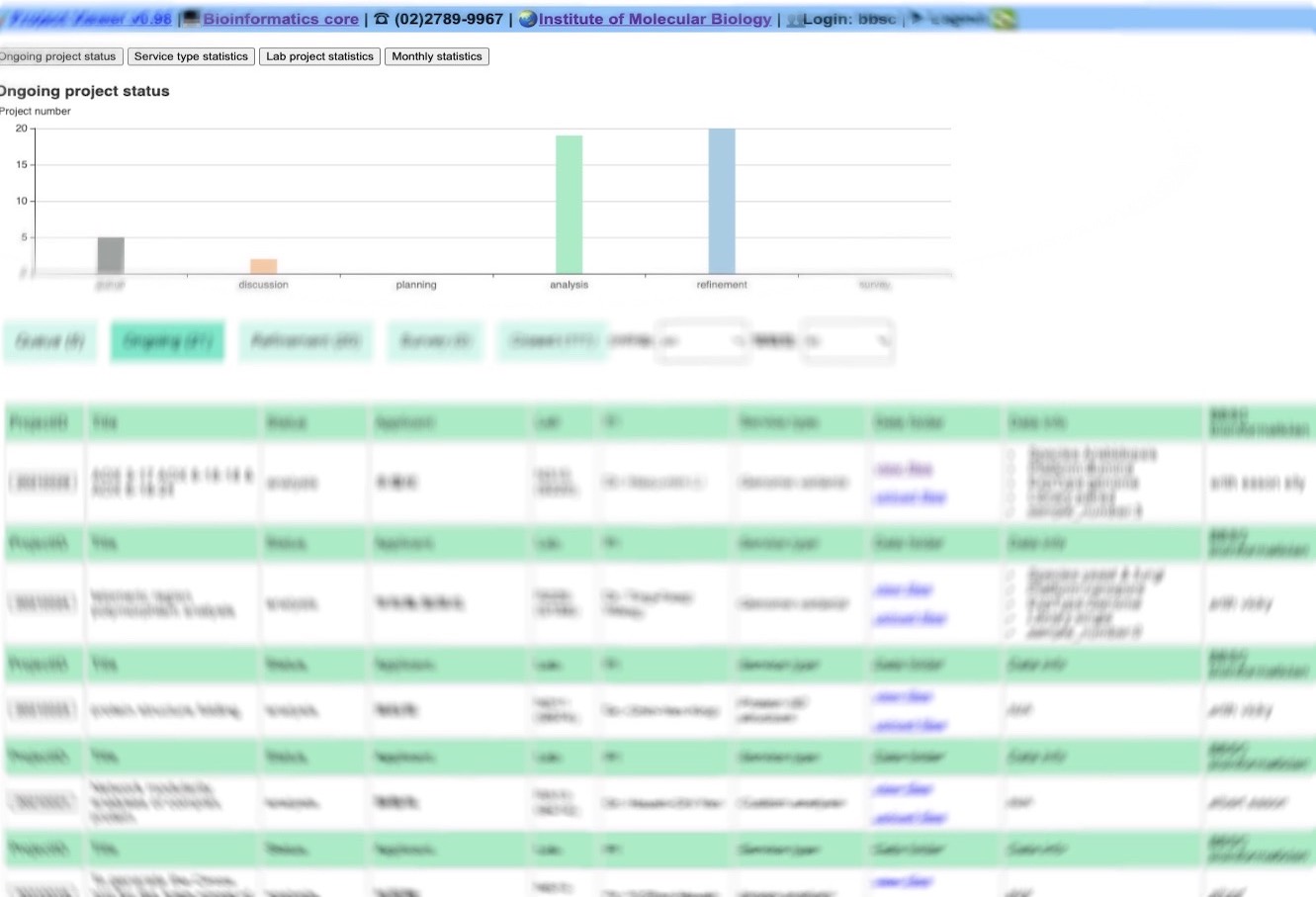
Project viewer is a user-friendly platform to upload data, track progress and discuss results. It is particular useful for viewing pasted projects.
Read More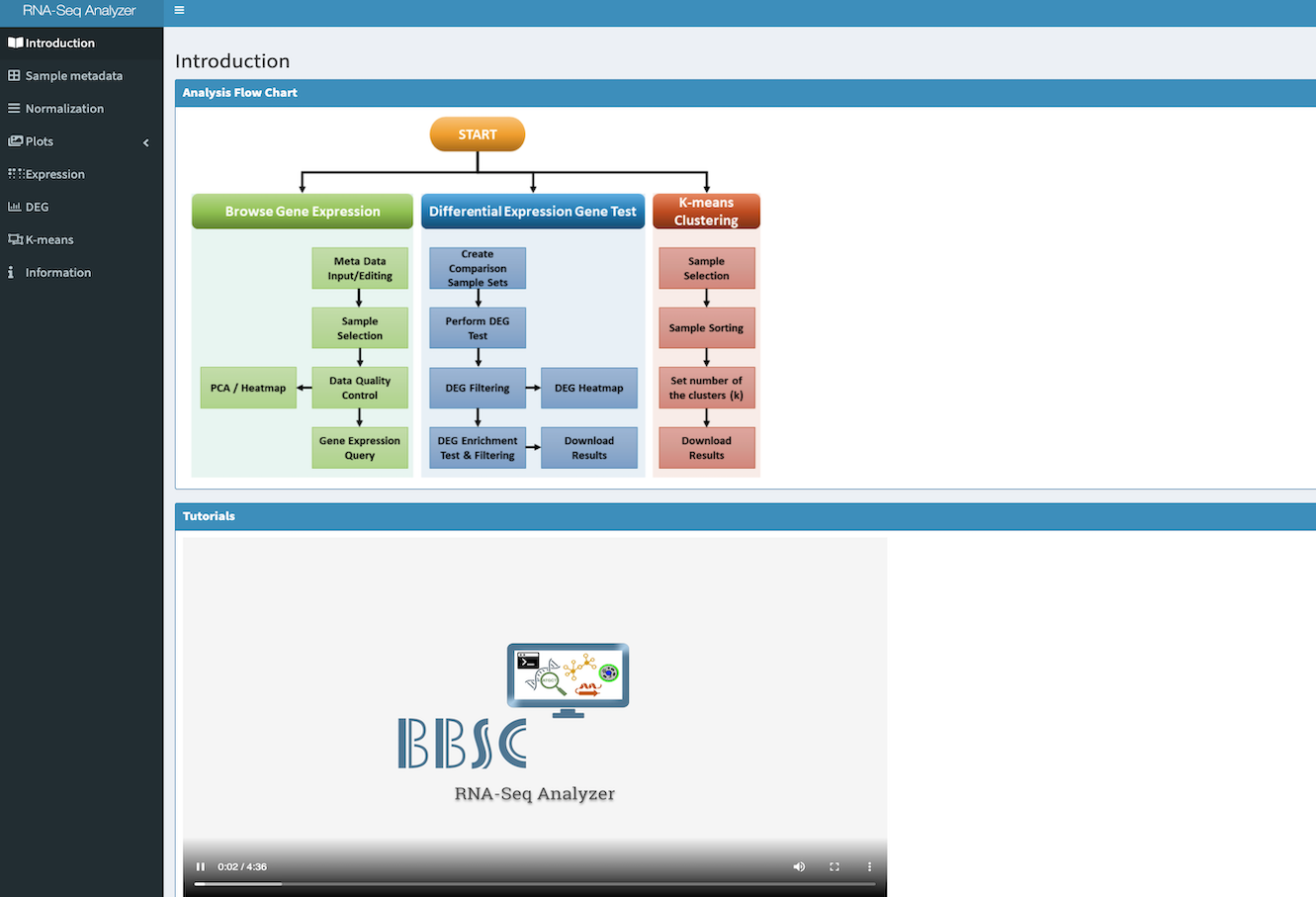
The cloud-based Bioinformatics apps for RNA-Seq analysis, variant filtering, and enrichment analysis.
Read More
余承欣 (Albert), Ph.D.
Chen-Hsin Yu
Core manager
(02) 2789-9967
albertchyu at as.edu.tw

林信男 (Arith), Ph.D.
Hsin-Nan Lin
Core Manager
(02)2789-9967
arith at as.edu.tw

葉坤海 (Eason), M.S.
Kun-Hai Yeh
Senior Bioinformatician
(02) 2789-9965
eason at as.edu.tw

董冠群 (Ken), M.S.
Kuan Chiun Tung
Bioinformatician
(02) 2789-9965
as0202185 at as.edu.tw
bsc@gate.sinica.edu.tw
(02) 2789-9965 / 2789-9967
Bioinformatics Core
Institute of Molecular Biology
Academia Sinica
Taipei, Taiwan